MBI Videos
Ben Wendel
-
 Ben Wendel
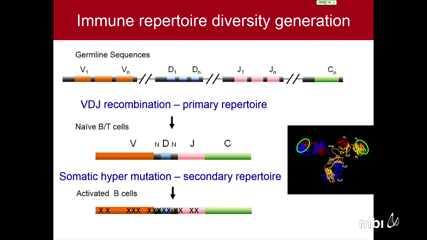
Ben WendelThe immune repertoire has incredible diversity generated through V(D)J recombination to recognize the universe of antigens. This diversity, while crucial for the immune system, makes immune repertoire sequencing (IR-seq) a challenging task. Current sequencing technologies lack the accuracy to delineate between the myriad of minor mutations and PCR/sequencing errors. Using a barcode-driven Molecular IDentifier clustering-based IR-Seq (MIDCIRS), we’ve effectively lowered the error rate for high throughput IR-seq to ~1 in 30,000 nucleotides. With this unprecedented accuracy, we have the power to resolve finer properties of the immune repertoire to characterize responses to disease, treatments, vaccinations, and aging.
On the other end of the spectrum, single cell analysis is critical to elucidate heterogeneity that can be lost in bulk samples. Antigen-specific T cells can be extremely rare, as low as 1 per million T cells, and can have starkly difference phenotypic and functional properties than the bulk. Using a pMHC tetramer-based enrichment strategy, we can isolate the responders to particular diseases and employ our single cell analysis method to simultaneously measure the T cell receptor sequences and gene expression levels. Our single cell analysis technique can be used to investigate the clonal nature and functional capacity of tumor infiltrating lymphocytes – the groups of T cells that are sought to activate by many cancer immunotherapies. This can lead to pretreatment screening and post-treatment disease monitoring methods that can be utilized to provide the optimal treatment regimen for a given patient.
